A self-assembling nanoparticle designed by a UConn professor is the key component of a potent new malaria vaccine that is showing promise in early tests.
For years, scientists trying to develop a malaria vaccine have been stymied by the malaria parasite’s ability to transform itself and “hide” in the liver and red blood cells of an infected person to avoid detection by the immune system.
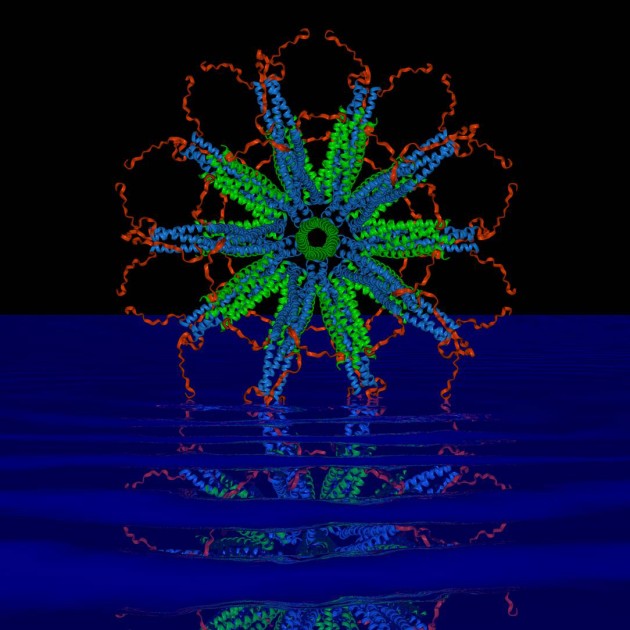
But a novel protein nanoparticle developed by Peter Burkhard, a professor in the Department of Molecular & Cell Biology, in collaboration with David Lanar, an infectious disease specialist with the Walter Reed Army Institute of Research, has shown to be effective at getting the immune system to attack the most lethal species of malaria parasite, Plasmodium falciparum, after it enters the body and before it has a chance to hide and aggressively spread.
The key to the vaccine’s success lies in the nanoparticle’s perfect icosahedral symmetry (think of the pattern on a soccer ball) and ability to carry on its surface up to 60 copies of the parasite’s protein. The proteins are arranged in a dense, carefully constructed cluster that the immune system perceives as a threat, prompting it to release large amounts of antibodies that can attack and kill the parasite.
In tests with mice, the vaccine was 90-100 percent effective in eradicating the Plasmodium falciparum parasite and maintaining long-term immunity over 15 months. That success rate is considerably higher than the reported success rate for RTS,S, the world’s most advanced malaria vaccine candidate currently undergoing phase 3 clinical trials, which is the last stage of testing before licensing.

“Both vaccines are similar, it’s just that the density of the RTS,S protein displays is much lower than ours,” says Burkhard. “The homogeneity of our vaccine is much higher, which produces a stronger immune system response. That is why we are confident that ours will be an improvement.
“Every single protein chain that forms our particle displays one of the pathogen’s protein molecules that are recognized by the immune system,” adds Burkhard, an expert in structural biology affiliated with UConn’s Institute of Materials Science. “With RTS,S, only about 14 percent of the vaccine’s protein is from the malaria parasite. We are able to achieve our high density because of the design of the nanoparticle, which we control.”
The research was published in Malaria Journal in 2013.
The search for a malaria vaccine is one of the most important research projects in global public health. The disease is commonly transported through the bites of nighttime mosquitoes. Those infected suffer from severe fevers, chills, and a flu-like illness. In severe cases, malaria causes seizures, severe anemia, respiratory distress, and kidney failure. Each year, more than 200 million cases of malaria are reported worldwide. The World Health Organization estimated that 627,000 people died from malaria in 2012, many of them children living in sub-Saharan Africa.
It took the researchers more than 10 years to finalize the precise assembly of the nanoparticle as the critical carrier of the vaccine and find the right parts of the malaria protein to trigger an effective immune response. The research was further complicated by the fact that the malaria parasite that impacts mice used in lab tests is structurally different from the one infecting humans.
The scientists used a creative approach to get around the problem.

“Testing the vaccine’s efficacy was difficult because the parasite that causes malaria in humans only grows in humans,” Lanar says. “But we developed a little trick. We took a mouse malaria parasite and put in its DNA a piece of DNA from the human malaria parasite that we wanted our vaccine to attack. That allowed us to conduct inexpensive mouse studies to test the vaccine before going to expensive human trials.”
The pair’s research has been supported by a $2 million grant from the National Institutes of Health and $2 million from the U.S. Military Infectious Disease Research Program. A request for an additional $7 million in funding from the U.S. Army to conduct the next phase of vaccine development, including manufacturing and human trials, is pending.
“We are on schedule to manufacture the vaccine for human use early next year,” says Lanar. “It will take about six months to finish quality control and toxicology studies on the final product and get permission from the FDA to do human trials.”
Lanar says the team hopes to begin early testing in humans in 2016 and, if the results are promising, field trials in malaria endemic areas will follow in 2017. The required field trial testing could last five years or more before the vaccine is available for licensure and public use, Lanar says.
Martin Edlund, CEO of Malaria No More, a New York-based nonprofit focused on fighting deaths from malaria, says, “This research presents a promising new approach to developing a malaria vaccine. Innovative work such as what’s being done at the University of Connecticut puts us closer than we’ve ever been to ending one of the world’s oldest, costliest, and deadliest diseases.”
A Switzerland-based company, Alpha-O-Peptides, founded by Burkhard, holds the patent on the self-assembling nanoparticle used in the malaria vaccine. Burkhard is also exploring other potential uses for the nanoparticle, including a vaccine that will fight animal flu and one that will help people with nicotine addiction. Professor Mazhar Khan from UConn’s Department of Pathobiology is collaborating with Burkhard on the animal flu vaccine.
article by Colin Poitras